Introduction:
Paediatric acute leukaemias are the most common form of childhood cancer. Mixed-lineage leukaemia (MLL) in particular is a very aggressive type of leukaemia in children. Although we have seen improvements in the identification of the different types of leukaemia and advances have been made in the range of treatments over recent years, the fact is that many patients suffer from a high-risk form and end up dying from the disease.
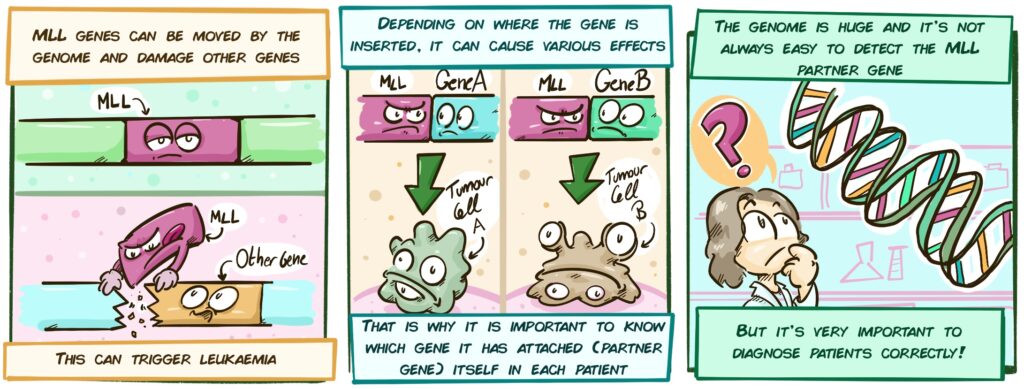
In order to be able to choose the best possible treatment, it is essential to identify as many characteristics of the cancer as possible in order to be able to classify patients based on their prognosis, i.e. rank them according to high or low risk of relapse. Other types of cancer are known for having a large number of highly diverse genetic abnormalities, which allows them to be classified and possible therapeutic targets identified. However, compared with other adult cancers, childhood leukaemias have very few DNA alterations. This is even more noticeable in so-called MLL. These leukaemias are characterised by the presence of a rearranged MLL gene (Mixed-Lineage Leukaemia, due to having the ability to hijack any cell line) and they are very common in babies (80% of infant leukaemias have this gene rearrangement).
In the case of MLL, the key lies not so much in the number of added mutations but rather in how this gene is altered or rearranged. What usually happens is that a fragment of this gene breaks off and becomes attached or fused to a different gene, which changes its function (these changes are called translocations). The genes to which MLL is attached or fused are called partner genes and they are the new travel companions of the MLL gene. Based on this partner gene, leukaemias behave differently, with varying degrees of aggressiveness and different prognoses. This is where there is a possibility to classify patients better and find more personalised treatment strategies.
The group led by Dr Camós and her team is leading the way in the study of MLL and the role played by biological alterations in both the prognosis and treatment of these children. Over the course of its work, this research group has focused on different aspects of these diseases in the form of two projects as outlined below.
Part 1 of the project: Epigenetic study of MLL
As we have already mentioned, unlike other types of cancer, childhood leukaemias have a low number of genetic alterations. There is, however, another type of modification that may perhaps explain the differences between patients and help us to understand and treat each patient better. We are talking about epigenetic alterations. Epigenetics refers to all of the processes involved in how, when and where the information contained in the DNA is read (not to the actual contents and structure of the DNA, which is what genetics studies). There are various molecules that regulate DNA reading, including:
- A group of proteins known as histone deacetylase (HDAC), which regulate which areas of the DNA are accessible for reading and which are not;
- Some very small fragments of genetic material known as microRNAs (miRNAs). Instruction books (known as RNA) are usually extracted from the DNA library to make proteins, which are actually responsible for performing cell functions. MicroRNAs are capable of preventing these instruction books from being used.
The combined study of miRNAs and HDACs may be highly relevant for several reasons:
- It allows us to gain a much deeper knowledge of this type of disease;
- It may allow us to identify more suitable biomarkers that allow us to predict patients’ outcome better;
- It will allow us to conduct new studies in the future to create more personalised therapies.
For this study, bone marrow samples taken from paediatric patients at several hospitals, including Hospital Sant Joan de Déu, Hospital Vall d’Hebrón and Hospital Universitario Niño Jesús de Madrid, have been used. Both babies and patients up to 18 years of age were included to study possible differences in the aggressiveness of this type of leukaemia in different age groups.
Part 2 of the project: Minimal residual disease analysis using new genetic techniques
One of the main challenges of childhood leukaemias is the correct diagnosis and follow-up of patients over time after administering treatments to see whether any tumour cells have been left behind that could result in a relapse (which is known as Measurable Residual Disease or MRD).
The diagnostic and follow-up methods used for the disease currently have some limitations:
- They are not able to accurately identify all MLL gene rearrangements and alterations and diagnoses may therefore be inaccurate;
- MRD follow-up is not sensitive and effective enough and the presence of tumour cells may not be detected in a high number of cases. This is a problem because relapses are detected too late and this entails a higher risk for the patient.
Therefore, new methods are required to improve both diagnosis and follow-up. The objective of this project is to use latest-generation techniques to detect and completely identify 100% of MLL gene rearrangements and to use more sensitive follow-up techniques that allow even very low levels of tumour cells to be detected.
[MLL genes can be moved by the genome and damage other genes
MLL
Other gene
This can trigger leukaemia
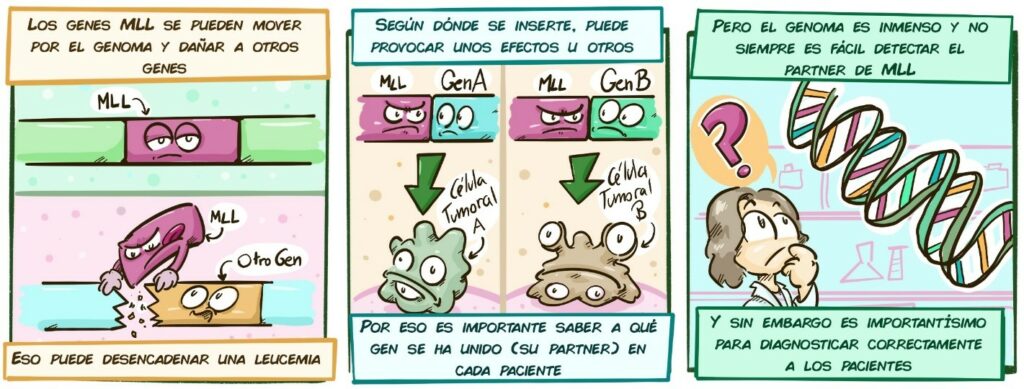
Depending on where the gene is inserted, it can cause various effects
MLL Gene A Gene B
Tumour cell A Tumour cell B
That is why it is important to know which gene it has attached itself to (its partner gene) in each patient But the genome is huge and it is not always easy to detect the MLL partner gene But this is very important to diagnose patients correctly]
Dr Camós’ group has proposed two different approaches:
- Targeted Locus Amplification (TLA):
- This is a special technique that makes it possible to identify any MLL gene modification, even modifications that have not been described before;
- The identification of changes in MLL may allow more precise and individualised follow-up with the specific detection of cells showing this change.
- Confirmation of MLL biomarkers:
- One study has documented that patients with certain MLL gene rearrangements have increased levels of certain genes (e.g. RET, PPP1R27, CCL23, SCUBE1 or PHACTR3).
- Increased activity of these genes at diagnosis could:
- be related to cell behaviour in some types of MLL;
- if these data are confirmed, we may be able to measure the activity of these genes to find out if the patient is doing better or worse.
Part 1 results:
Several interesting findings have been reported in the HDAC study. Firstly, it has been observed that two proteins (HDAC9 and MEF2D) are detected more in patients with this type of leukaemia, regardless of age and lineage (by lineage we are referring to whether the patient has myeloid or lymphoid leukaemia). Also, an increase in HDAC9 levels has been associated with poorer prognosis.
These results may play an important role in the development of future targeted therapies. Future treatments against these HDACs may be used to support current therapies in order to increase their effectiveness.
In the miRNA study, despite the pandemic, the team has managed to make great progress. It has already managed to analyse more than 90 different microRNAs in more than 100 patients. In fact, a huge amount of preliminary data has been generated, showing that, according to the characteristics of each patient’s leukaemia, some miRNAs are increased while others are decreased. This again opens the way for future more personalised treatments.
Combining the miRNA and HDAC data with patients’ genetic and follow-up data has resulted in an extensive database and data are currently being analysed as they have the potential to provide highly relevant conclusions.
Part 2 results:
With regards to the biomarker study, the group’s results are very preliminary but they appear to confirm that patients with some MLL gene rearrangements have higher levels of those genes studied. It is still necessary to analyse more patients to be able to certify these results and confirm the specificity and possible usefulness of these biomarkers for patient follow-up in hospital practice.
As far as the analysis of patients using the TLA technique is concerned, Dr Camós’ team has analysed samples from 32 patients with MLL. In the majority of these samples, the team managed to accurately identify where the MLL gene had moved. However, the TLA technique did not manage to identify the specific rearrangement in some patients.
Therefore, although the techniques used to date to identify genetic changes in each patient’s tumour work fairly well, there are still a few cases where such identification is not possible. Nevertheless, it is vital to be able to identify as close to 100% of the patients with MLL gene rearrangements in order to achieve more personalised medicine.
This is why Dr Camós’ team has developed an innovative strategy involving a work flow to apply different diagnostic techniques for each patient with MLL gene rearrangements.
- Firstly, patients will be initially analysed to look for MLL gene rearrangements using conventional techniques already used for diagnosis.
- For a more detailed study of MLL patients:
- TLA will be used in those patients from whom a sample of sufficient quality has been taken;
- If TLA does not identify the precise alteration, or if the sample is not large enough or of an adequate quality, other techniques (such as RNA-Seq) will be used to confirm or rule out the presence of MLL gene alterations.
- Once each child’s specific alteration has been identified, targeted detection probes can be designed to identify the tumour cells of that specific patient in a personalised manner. This allows the use of more specific and precise techniques to check whether tumour cells are still present, have been eliminated or are increasing.
Overall, the aim of this strategy is to identify, as precisely as possible, 100% of patients with MLL gene rearrangements and to achieve more sensitive and effective follow-up of the patient’s disease. This will improve therapeutic decisions, improve reaction time against potential relapses and finally improve patient survival and quality of life.